The work, from the Wellcome Sanger Institute, the French Institute of Health and Medical Research (Inserm), the University of Bourgogne Franche-Comté, and Beijing Genomics Institute (BGI)-Shenzhen, found that when there was a loss-of-function mutation in the mouse gene Magee2, it led to adult male mice having slightly enlarged brains. When the researchers investigated human brains using scans, they found the same effects in people who have a naturally-occurring loss-of-function mutation in the corresponding human gene, MAGEE2.
The paper, published in Molecular Biology and Evolution, provides an example of beneficial, positive selection of a mutated gene, in this case where the loss of function could provide potential advantages.
This MAGEE2 variant, found mainly in East Asia and the Americas, adds to the current genetic understanding of evolution and is an example of local adaptation, even though the exact reason behind this selection is so far unknown. More studies are needed to fully understand the role of MAGEE2 when it is intact and the impact, if any, of its loss on cognitive function.
Gene inactivation, where a gene is mutated and therefore no longer works, is often considered harmful and linked to the development of disease. However, if the gene that is lost isn’t responsible for an essential function, the mutation could have no impact on the individual, or in some cases might provide evolutionary advantages. This has been documented previously with some mutations being linked to malaria resistance*, or norovirus resistance**.
In earlier work, the mutated MAGEE2 gene was identified as one of the strongest examples of positive selection in East Asia, being found at 84 per cent frequency across the region and in the Americas, but only at low frequency elsewhere. While this has been known for some time, there is little understanding about the function of the gene or the reasons behind its selection.***
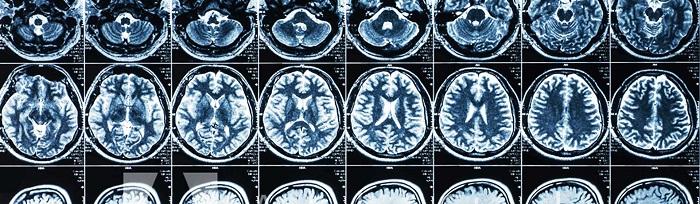
The new research, from the Wellcome Sanger Institute and collaborators, used CRISPR-Cas9 genome editing to create knockout mouse models to investigate the impact of MAGEE2 inactivation, and then compare these findings to humans by analysing the brain scans of those who carry the inactivated gene.
They found that in mice, inactivation of Magee2 led to an increase of 13 per cent in size of the brain in male mice, with no obvious impact on health or behaviour. This was mimicked in humans, where MRI brain scans showed an enlarged brain in men with an inactivated MAGEE2. In humans, there was a decrease in brain size in women with an inactivated MAGEE2, a trend also seen in mice, although not statistically significant.
Further studies are now needed to understand the function of the gene, and the full impact of its loss on cognition and behaviour.
Dr Michal Szpak, first author and Postdoctoral Fellow at the Wellcome Sanger Institute, said: “MAGEE2 inactivation has been recognised by its striking geographical distribution, suggestive of a strong positive selection in East Asia, yet with no understanding of its function. This is, to our knowledge, the first study that links the inactivation of MAGEE2 in humans and mice to the enlargement of brain structures. We now need to investigate this further to fully understand whether this variant affects cognition or behaviour, what the evolutionary advantage is, and why we see opposite effects in men and women.”
Dr Binnaz Yalcin, senior author and Junior Group Leader at the French Institute of Health and Medical Research (Inserm), said: “This study is a powerful example of how the mouse remains crucial as a model system beyond the field of medical genetics. We are pleased to see how findings in mice can translate to the biology of human genome evolution.”
Professor Huanming Yang, Co-founder, director and chairman of the Beijing Genomics Institute Group, China, said: “Understanding the selection of genes in different populations can help build a more complete picture of genetic evolution and lead to a deeper understanding of the role of non-essential genes in the body. Investigating these non-essential genes and their impact on brain size could lead to insight on what drove the genetic changes in this group and the role of different genes on the function of the brain. The more we understand about the brain, the better we will be at being able to understand and treat brain diseases that are caused by genetic mutations.”
Dr Chris Tyler-Smith, senior author and former Senior Group Leader at the Wellcome Sanger Institute, said: “We could tell from previous genetic studies of MAGEE2 in the population that its loss was beneficial, at least in some parts of the world, but we had no idea why. Now we know from the current work starting in mice that loss has effects on brain size. In future research we need to go back to the human population to see how these brain changes, or others associated with MAGEE2 loss, have led to the beneficial effects that started off this work.”
Image credit: AdobeStock
* Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki A. (2005). Evolution of human-chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proc Natl Acad Sci U S A 102:12819-12824.
** Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. (1995). Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem 270:4640-4649.
*** Yngvadottir B, Xue Y, Searle S, Hunt S, Delgado M, Morrison J, Whittaker P, Deloukas P, Tyler-Smith C. (2009). A genome-wide survey of the prevalence and evolutionary forces acting on human nonsense SNPs. Am J Hum Genet 84:224-234.
*** Szpak M, Mezzavilla M, Ayub Q, Chen Y, Xue Y, Tyler-Smith C. (2018). FineMAV: prioritizing candidate genetic variants driving local adaptations in human populations. Genome Biol 19:5.
Publication:
M. Szpak, S. Collins, Y. Li, X. Liu, et al. (2021) A positively-selected MAGEE2 LoF allele is associated with sexual dimorphism in human brain size, and shows similar phenotypes in Magee2 null mice. Molecular Biology and Evolution. DOI: 10.1093/molbev/msab243